Introducing Distributed LightGBM Training with Ray
LightGBM is a gradient boosting framework based on tree-based learning algorithms. Compared to XGBoost, it is a relatively new framework, but one that is quickly becoming popular in both academic and production use cases.
Today, we’re excited to announce a beta release of LightGBM on Ray, which lets you easily distribute your training on your Ray cluster.
Features of LightGBM-Ray include:
Multi-node and multi-GPU training support
Seamless integration with Ray Tune for distributed hyperparameter search
LinkWhy yet another gradient boosting algorithm?
If you are familiar with gradient boosting methods, you are likely well-versed with XGBoost, which has established itself as the standard for training ML models on tabular data. XGBoost is a terrific library that is used extensively in production by ML-heavyweights like Uber and Stripe.
So you might be wondering, why yet another gradient boosting library? LightGBM is designed to be fast and efficient, and offers several advantages:
Faster training
Built-in support for categorical variables
Optimizations for training on larger datasets
Better accuracy in certain situations
The good news is that you don’t have to choose. Since it is based on XGBoost-Ray, the LightGBM on Ray integration lets you easily switch between XGBoost on Ray and LightGBM on Ray for your classification or regression problems (support for ranking coming soon), compare the results, and choose the one that works best for your usecase.
LinkGetting started with LightGBM on Ray
Let’s take a look at a few examples that show you can easily port your existing LightGBM code to use this new integration, and run in distributed mode. In order to run all the code in this section, scikit-learn and lightgbm_ray need to be installed. This can be done with
1pip install sklearn lightgbm_rayLinkTraining
Let’s start with a simple, non-distributed example, running on a single node with core LightGBM.
1from lightgbm import Dataset, train
2from sklearn.datasets import load_breast_cancer
3
4train_x, train_y = load_breast_cancer(return_X_y=True)
5train_set = Dataset(train_x, train_y)
6
7evals_result = {}
8bst = train(
9 {
10 "objective": "binary",
11 "metric": ["binary_logloss", "binary_error"],
12 },
13 train_set,
14 num_boost_round=10,
15 evals_result=evals_result,
16 valid_sets=[train_set],
17 valid_names=["train"],
18 verbose_eval=False,)
19
20bst.save_model("model.lgbm")If your run is successful, you should see a number of [LightGBM] [Warning] No further splits with positive gain, best gain: -inf lines as LightGBM finishes training and decides no more splits are needed.
Now let’s scale out. Just by changing four lines of code, we can turn this into a distributed run. The changes have been highlighted below.
1from lightgbm_ray import RayDMatrix, RayParams, train
2from sklearn.datasets import load_breast_cancer
3
4train_x, train_y = load_breast_cancer(return_X_y=True)
5train_set = RayDMatrix(train_x, train_y)
6
7evals_result = {}
8bst = train(
9 {
10 "objective": "binary",
11 "metric": ["binary_logloss", "binary_error"],
12 },
13 train_set,
14 num_boost_round=10,
15 evals_result=evals_result,
16 valid_sets=[train_set],
17 valid_names=["train"],
18 verbose_eval=False,
19 ray_params=RayParams(num_actors=2, cpus_per_actor=2))
20
21bst.booster_.save_model("model.lgbm")Like XGBoost-Ray, LightGBM-Ray also uses the RayDMatrix class for datasets. This lets you easily switch between LightGBM-Ray and XGBoost-Ray just by changing the import statement.
LinkPrediction
Distributed inference can be run in a similar way to XGBoost-Ray.
1from sklearn import datasets
2import lightgbm as lgbm
3from lightgbm_ray import RayDMatrix, predict
4
5data, labels = datasets.load_breast_cancer(return_X_y=True)
6
7dpred = RayDMatrix(data, labels)
8
9bst = lgbm.Booster(model_file="model.lgbm")
10predictions = predict(bst, dpred)
11
12print(predictions)LinkScikit-learn API
LightGBM-Ray also provides a fully functional scikit-learn API for both training and prediction. You can either use NumPy arrays or pandas dataframes, which will be converted internally, or you can pass a RayDMatrix object for greater control.
1from lightgbm_ray import RayLGBMClassifier, RayParams
2from sklearn.datasets import load_breast_cancer
3from sklearn.model_selection import train_test_split
4
5seed = 42
6
7X, y = load_breast_cancer(return_X_y=True)
8X_train, X_test, y_train, y_test = train_test_split(
9 X, y, train_size=0.25, random_state=42)
10
11clf = RayLGBMClassifier(
12 n_jobs=2, # In LightGBM-Ray, n_jobs sets the number of actors
13 random_state=seed)
14
15# scikit-learn API will automatically convert the data to RayDMatrix format as needed.
16# You can also pass X as a RayDMatrix, in which case y will be ignored.
17
18clf.fit(X_train, y_train)
19
20pred_ray = clf.predict(X_test)
21print(pred_ray)
22
23pred_proba_ray = clf.predict_proba(X_test)
24print(pred_proba_ray)
25
26# It is also possible to pass a RayParams object to fit/predict/predict_proba methods - will override
27# n_jobs set during initialization
28
29clf.fit(X_train, y_train, ray_params=RayParams(num_actors=2))
30
31pred_ray = clf.predict(X_test, ray_params=RayParams(num_actors=2))
32print(pred_ray)LinkOK, but how do I load real-world data into LightGBM-Ray?
See the Higgs LightGBM-Ray example code from the Github repository.
There you’ll find a full example that reads from a CSV and trains a classification model.
LinkBenchmarking
We have run two simple benchmarks comparing LightGBM-Ray to XGBoost-Ray and non-distributed LightGBM.
Comparison to XGBoost depending on the number of workers
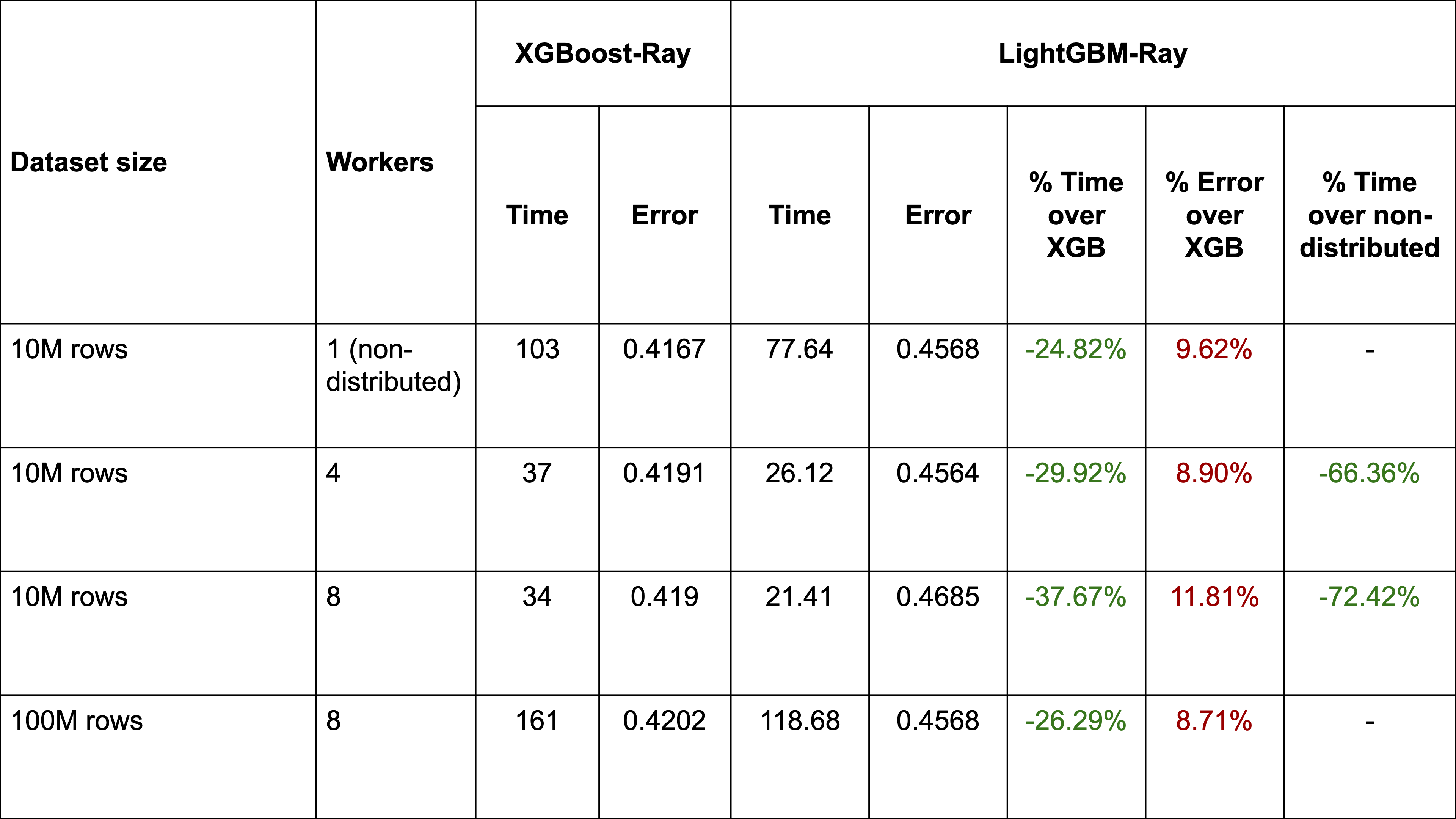
We can see that with a large synthetic dataset, distributing LightGBM using Ray can reduce training time by over 66%. Furthermore, LightGBM-Ray consistently outperforms XGBoost-Ray on training time, but does lose out on accuracy (for this particular dataset).
Comparison with XGBoost-Ray during hyperparameter tuning with Ray Tune
This experiment was conducted using a million row dataset and a 75-25 train-test split. Both XGBoost-Ray and LightGBM-Ray were distributed over 8 actors per trial, each using 2 threads. There were 4 trials running concurrently with a deadline of 5 minutes. The search spaces were identical.
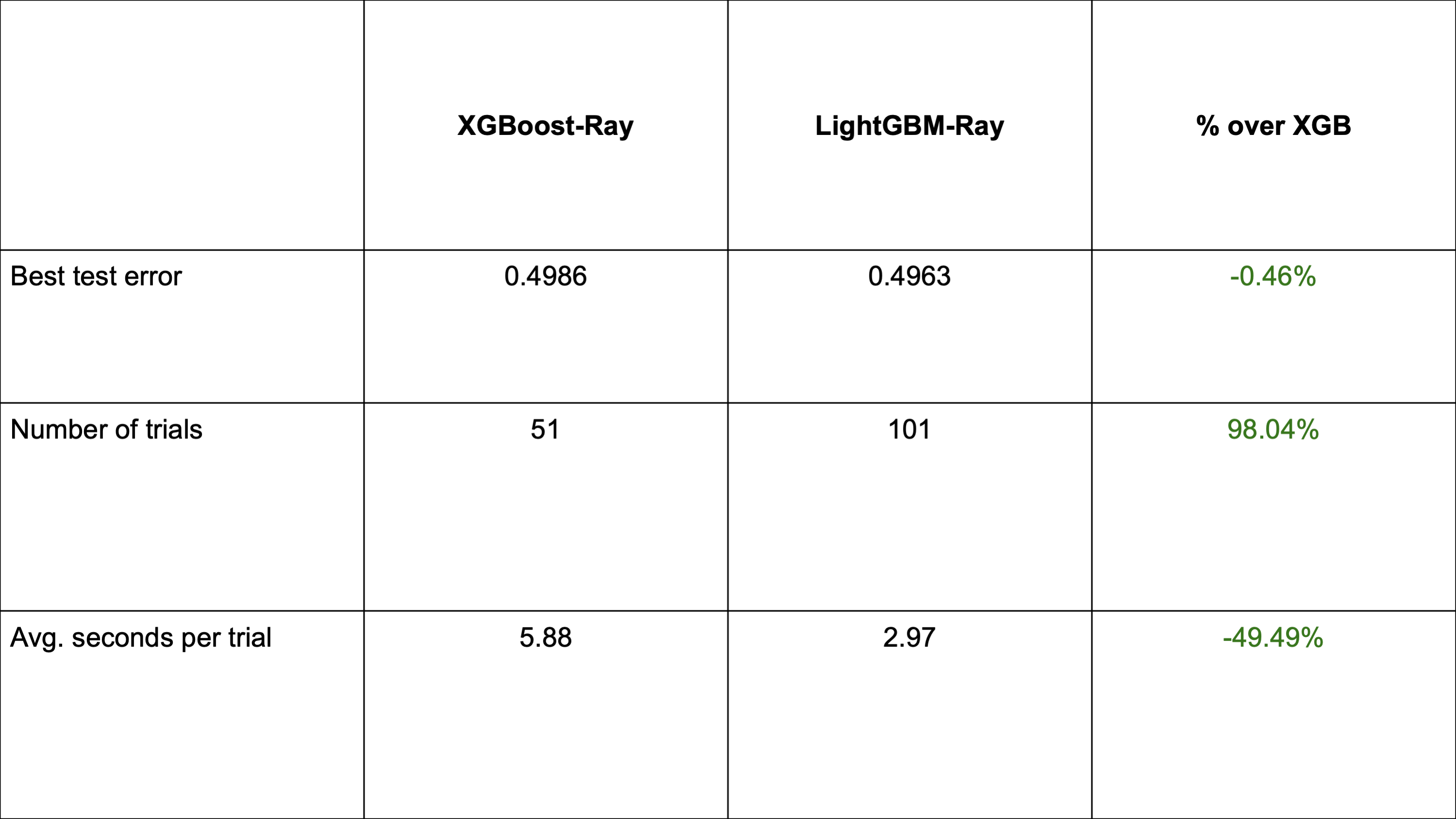
By using Ray Tune for hyperparameter optimization, LightGBM is able to narrow the gap between XGBoost-Ray in terms of accuracy, even slightly outperforming it in the same time budget. As LightGBM trains faster than XGBoost, it is possible to evaluate more hyperparameter combinations, increasing the chances of finding those that increase accuracy.
LinkArchitecture of LightGBM-Ray
LightGBM-Ray does not change how LightGBM works. Instead, it manages the data sharding and actors through Ray. It distributes LightGBM training and prediction by dividing up the data among several Ray Actors, running either on your laptop or in a multi-node Ray cluster. Each of those Actors then uses built-in LightGBM socket-based communication to share information about the training state.
Furthermore, LightGBM-Ray’s fault tolerance mechanisms ensure that training will be automatically restarted (without the need to read data again) should an Actor die for any reason.
This is a very similar setup to XGBoost-Ray.
LinkKnown Issues
As of now, due to many common internals, LightGBM-Ray uses XGBoost-Ray as a hard dependency, necessitating XGBoost to be installed. We are working on removing that requirement. Furthermore, elastic training (present in XGBoost-Ray) is not yet supported, but will be added in a future release.
Using too many actors (controlled by the num_actors parameter in a RayParams object) may result in a drop of accuracy, if each of them receives too small of a portion of a dataset. The same issue may occur if the data isn’t well-shuffled. This is due to how LightGBM conducts distributed training. Additionally, in order to ensure efficient training, each actor requires at least two CPUs, so that the communication thread can run without blocking (as per LightGBM documentation).
LinkConclusion
LightGBM is a fast training and accurate alternative to XGBoost that offers many advantages. With LightGBM on Ray, it’s now possible to scale your LightGBM code on any cloud provider with just a few code changes .
We welcome feedback and issues on GitHub and on the Ray Discourse. Let us know if you run into problems!


